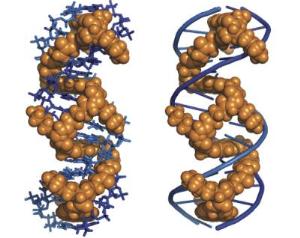
AUSTIN, Texas - Chemists at The University of Texas at Austin have created a molecule that's so good at tangling itself inside the double helix of a DNA sequence that it can stay there for up to 16 days before the DNA liberates itself, much longer than any other molecule reported.
It's an important step along the path to someday creating drugs that can go after rogue DNA directly.
Such drugs would be revolutionary in the treatment of genetic diseases, cancer or retroviruses such as HIV, which incorporate viral DNA directly into the body's DNA.
"If you think of DNA as a spiral staircase," says Brent Iverson, professor of chemistry and chair of the department of chemistry and biochemistry, "imagine sliding something between the steps. That's what our molecule does. It can be visualized as binding to DNA in the same way a snake might climb a ladder. It goes back and forth through the central staircase with sections of it between the steps. Once in, it takes a long time to get loose. Our off-rate under the conditions we used is the slowest we know of by a wide margin."
Iverson says the goal is to be able to directly turn on or off a particular sequence of genes.
"Take HIV, for example," he says. "We want to be able to track it to wherever it is in the chromosome and just sit on it and keep it quiet. Right now we treat HIV at a much later stage with drugs such as the protease inhibitors, but at the end of the day, the HIV DNA is still there. This would be a way to silence that stuff at its source."
Iverson, whose results were published in Nature Chemistry and presented this month at a colloquium at NYU, strongly cautions that there are numerous obstacles to overcome before such treatments could become available.
The hypothetical drug would have to be able to get into cells and hunt down a long and specific DNA sequence in the right region of our genome. It would have to be able to bind to that sequence and stay there long enough to be therapeutically meaningful.
"Those are the big hurdles, but we jumped over two of them," says Iverson. "I'll give presentations in which I begin by asking: Can DNA be a highly specific drug target? When I start, a lot of the scientists in the audience think it's a ridiculous question. By the time I'm done, and I've shown them what we can do, it's not so ridiculous anymore."
In order to synthesize their binding molecule, Iverson and his colleagues begin with the base molecule naphthalenetetracarboxylic diimide (NDI). It's a molecule that Iverson's lab has been studying for more than a decade.
They then piece NDI units together like a chain of tinker toys.
"It's pretty simple for us to make," says Amy Rhoden Smith, a doctoral student in Iverson's lab and co-author on the paper. "We are able to grow the chain of NDIs from special resin beads. We run reactions right on the beads, attach pieces in the proper order and keep growing the molecules until we are ready to cleave them off. It's mostly automated at this point."
Rhoden Smith says that the modular nature of these NDI chains, and the ease of assembly, should help enormously as they work toward developing molecules that bind to longer and more biologically significant DNA sequences.
"The larger molecule is composed of little pieces that bind to short segments of DNA, kind of like the way Legos fit together," she says. "The little pieces can bind different sequences, and we can put them together in different ways. We can put the Legos in a different arrangement. Then we scan for sequences that they'll bind."
Iverson and Rhoden Smith's co-authors on the paper were Maha Zewail-Foote, a visiting scientist in Iverson's lab who's now an associate professor and chairman of chemistry at Southwestern University in Georgetown; Garen Holman, another former doctoral student of Iverson's who did most of the experimental work before obtaining his Ph.D.; and Kenneth Johnson, the Roger J. Williams Centennial Professor in Biochemistry at The University of Texas at Austin.
Further Information:
Garen G. Holman, Maha Zewail-Foote, Amy Rhoden Smith, Kenneth A. Johnson and Brent L. Iverson:
A sequence-specific threading tetra-intercalator with an extremely slow dissociation rate constant.
In: Nature Chemistry; 3, 875 - 881, 25 September 2011, DOI 10.1038/nchem.1151
Source: University of Texas at Austin, USA
Last update: 14.02.2012
Perma link: https://www.internetchemistry.com/news/2012/feb12/dna-tetra-intercalator.php
More chemistry: index | chemicals | lab equipment | job vacancies | sitemap
Internetchemistry: home | about | contact | imprint | privacy
© 1996 - 2023 Internetchemistry
