RUB-Researchers from the Chair for Biophysics have developed a new method for the detailed study of the interaction between pharmaceuticals and their target proteins.
The pharmaceutical industry has already taken notice of the new infrared spectroscopy technique; the method is supposed to be implemented to investigate pharmacological agent-protein interactions in the EU project K4DD, which is supported by various major European pharmaceutical companies.
"We now have a tool in our hands with which we can research the dynamics of pharmacologically interesting proteins in atomic detail," Prof. Dr. Klaus Gerwert said. "We want to undertake a targeted screening of substance libraries to look for potential pharmacological agents." PD Dr. Carsten Kötting added that "with our technique future pharmaceuticals can be more closely tailored to illness-causing proteins, which can noticeably reduce the negative side effects of these drugs."
They described the new method together with Dr. Jörn Güldenhaupt and Philipp Pinkerneil in the scientific journal 'ChemPhysChem' [see below], which dedicated its cover story to this topic.
The new method: from three to one
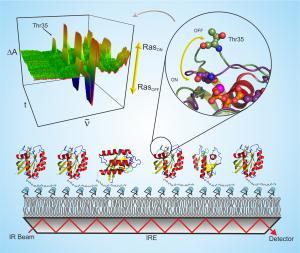
With infrared difference spectroscopy, researchers follow dynamic processes in proteins. For a long time, these processes could only be observed in light-activated proteins, but not in proteins that are activated by binding with ligands - but this is usually how many illness relevant molecules are activated. To analyze the dynamics of such proteins, researchers have to fasten them to the measurement surface and pour a pharmacological-substance over them; the proteins can then interact with and be activated by this substance. Even though this binding technique is possible, it cannot be used for all proteins. The RUB-Team worked around this problem by combining infrared (IR) spectroscopy with a surface-sensitive technique (attenuated total reflectance) and so-called "His-Tagging" (anchoring proteins to the measurement surface).
Attenuated total reflectance: bringing the infrared beam to all proteins
In conventional IR spectroscopy, an infrared beam is passed through a liquid sample; part of the light is absorbed by the proteins, which allows researchers to draw conclusions about their structure. The RUB-researchers beamed the infrared light through a germanium crystal, on whose surface proteins were anchored. At the boundaries of the crystal the light is reflected over and over, thereby spreading throughout the crystal (attenuated total reflectance). During this process, some of the light waves leave the crystal and reach the proteins that are fastened to its surface. A similar technique, the Surface Plasmon Resonance, is the standard for use in the pharmaceutical industry, but does not have the atomic resolution capabilities of the new technique.
Part of the chain: the His-Tag
This bonding of the proteins to the crystal succeeds through usage of the His-Tag, a simple amino acid chain, which is commonly attached to proteins today to enable their biochemical study - it is essentially a universal adapter. Through the His-Tag the RUB-researchers were able to anchor the protein to the germanium crystal. As a result the molecules were firmly bound to the measurement surface, which transmits the infrared light to the proteins by the process of attenuated total reflectance. The big advantage: an abundance of proteins are already fitted with the His-Tag; therefore examining them with the new method is unproblematic. All other proteins to which a His-Tag is attached can now also be accessed by IR spectroscopy. "This will help answer a multitude of biological and medical questions," Gerwert said.
Establishment of the new method with the switch protein Ras
The RUB-Team first tried their new method on the switch protein Ras, the central on/off switch for cell growth. Defect, or oncogenic Ras, is one of the cells most frequently responsible for causing cancer. The researchers succeeded in fastening Ras to the measurement surface with the His-Tag, and then activating the Ras by binding it to a ligand. "The technique is so sensitive that we could resolve the signal of a five nanometer thick protein layer. That"s about 1/10000 of the diameter of a human hair," RUB-researcher Dr. Jörn Güldenhaupt, who contributed significantly to the development of the new method, said. Even the smallest structural changes during the Ras protein"s switch from its "on" to its "off" state were recognized with the "protein-nanoscope."
Project funding
Funding for the project came from the Protein Research Department at the RUB, from the state of NRW in the framework of the Center for Vibrational Microscopy (CVM) and from the SFB 642, "GTP and ATP Dependent Membrane Processes," whose speaker is Prof. Gerwert.
Further Information:
Philipp Pinkerneil, Dr. Jorn Guldenhaupt, Prof. Dr. Klaus Gerwert, Dr. habil. Carsten Kotting:
Surface-Attached Polyhistidine-Tag Proteins Characterized by FTIR Difference Spectroscopy.
In: ChemPhysChem; Volume 13, Issue 11, pages 2649 - 2653, 06 August 2012, DOI 10.1002/cphc.201200358
Source: Ruhr University, Bochum, RUB, Germany
Last update: 05.09.2012
Perma link: https://www.internetchemistry.com/news/2012/sep12/anchored-proteins.php
More chemistry: index | chemicals | lab equipment | job vacancies | sitemap
Internetchemistry: home | about | contact | imprint | privacy
© 1996 - 2023 Internetchemistry
